This transformative framework taps into neural network learning patterns to correct ambiguous annotations, uncover hidden cell states, and enhance our understanding of health and disease.
 Research: Interpreting single-cell and spatial omics data using deep neural network training dynamics. Image Credit: Sergei Drozd / Shutterstock
Research: Interpreting single-cell and spatial omics data using deep neural network training dynamics. Image Credit: Sergei Drozd / Shutterstock
Annotatability is a robust new framework to address a significant challenge in biological research by examining how artificial neural networks (ANNs) learn to label genomic data. Genomic datasets often contain vast amounts of annotated samples, but many of these samples are annotated either incorrectly or ambiguously.
Borrowing from recent advances in the fields of natural language processing and computer vision, the team used artificial neural networks (ANNs) in a non-conventional way. Instead of merely using the ANNs to make predictions, the group inspected the difficulty with which they learned to label different biological samples. The difficulty was quantified using two key metrics: confidence and variability scores, which measure how well the neural network matches annotations to biological data.
Similar to assessing why students find some examples harder than others, the team then leveraged this unique source of information to identify mismatches in cell annotations, improve data interpretation, and uncover key cellular pathways linked to development and disease. This approach also highlights intermediate cell states, offering insights into the continuous nature of cellular development and differentiation. Annotatability provides a more accurate method for analyzing genomic data on single cells, offering significant potential for advancing biological research and, in the longer term, improving disease diagnosis and treatment.
A new study led by Jonathan Karin, Reshef Mintz, Dr. Barak Raveh, and Dr. Mor Nitzan from Hebrew University, published in the journal Nature Computational Science, introduces a new framework for interpreting single-cell and spatial omics data by monitoring deep neural networks’ training dynamics. The research aims to address the inherent ambiguities in cell annotations and offers a novel approach to understanding complex biological data.
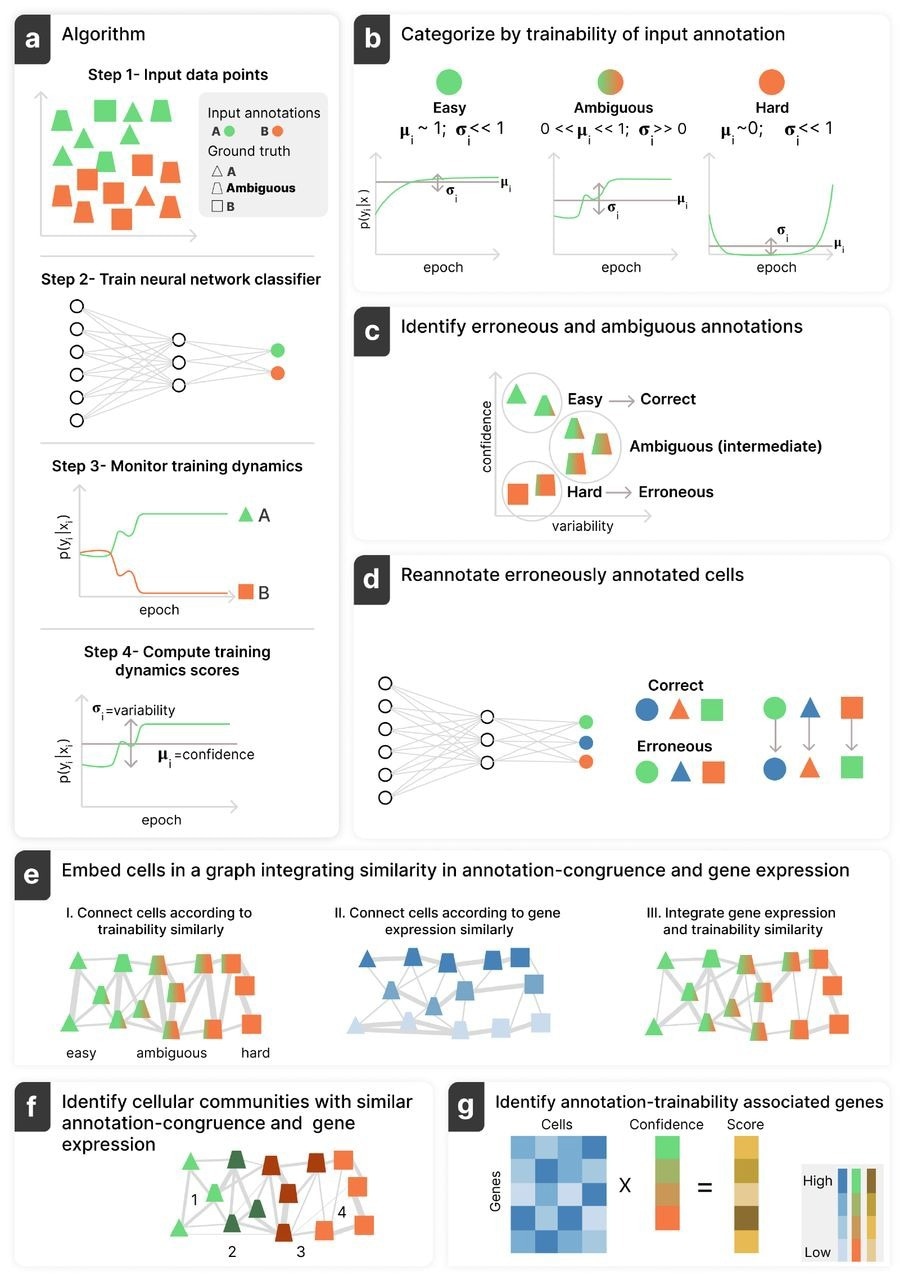
Annotatability schematic workflow. (a) Given an annotated dataset consisting of observations (e.g. single-cell gene expression profiles) and corresponding annotations per cell (e.g. cell types) (Step 1), Annotatability trains a DNN to predict the input annotations (Step 2), analyzes its predictions along the training procedure (Step 3), and computes confidence and variability scores per cell (Step 4). (b, c) Each cell is subsequently classified as easy-to-learn (high confidence/low variability), hard-to-learn (low confidence/low variability), or ambiguous (mid confidence/high variability)(b), corresponding to correctly annotated, erroneously annotated, or ambiguously annotated cells, respectively (c). (d) Annotatability can potentially amend the annotations of cells identified as erroneously annotated using its re-annotation module. (e, f) Annotatability includes a trainability-aware graph-embedding module, which incorporates similarity between cells in both training dynamics statistics as well as gene expression (e), enabling signal-specific downstream analyses (f). (g) Annotatability is also equipped with a training-dynamics-based score that captures either positive or negative association of genes relative to a given annotations, corresponding to a given biological signal. Image Credit: The Authors / https://www.biorxiv.org/
Single-cell and spatial omics data have transformed our ability to explore cellular diversity and cellular behaviors in health and disease. However, the interpretation of these high-dimensional datasets is challenging, primarily due to the difficulty of assigning discrete and accurate annotations, such as cell types or states, to heterogeneous cell populations. These annotations are often subjective, noisy, and incomplete, making it challenging to extract meaningful insights from the data.
The researchers developed a new framework, Annotatability, which helps identify mismatches in cell annotations and better characterizes biological data structures. By monitoring the dynamics and difficulty of training a deep neural network over annotated data, Annotatability identifies areas where cell annotations are ambiguous or erroneous. The framework is particularly effective at pinpointing areas where cells deviate from their expected behavior, as measured by their training dynamics.
As part of the study, the team introduced a signal-aware graph embedding method that enables more precise downstream analysis of biological signals. This technique captures cellular communities associated with target signals and facilitates the exploration of cellular heterogeneity, developmental pathways, and disease trajectories. Unlike traditional methods, this embedding approach integrates both gene expression data and annotation confidence to construct a more nuanced representation of biological variation.
The study demonstrates the applicability of Annotatability across a range of single-cell RNA sequencing and spatial omics datasets. Notable findings include the identification of erroneous annotations, delineation of developmental and disease-related cell states, and better characterization of cellular heterogeneity. For example, in spatial transcriptomics datasets, Annotatability was used to reannotate mislabeled cells, significantly improving data quality without discarding valuable spatial information. The results highlight the potential of this framework for unraveling complex cellular behaviors and advancing our understanding of both health and disease at the single-cell level.
The researchers' work presents a significant step forward in genomic data interpretation, offering a powerful tool for unraveling cellular diversity and enhancing our ability to study the dynamics of health and disease. Future developments of Annotatability aim to address challenges such as handling aggregated data, refining hyperparameter tuning, and enabling the analysis of continuous annotations. These advancements promise to broaden its applicability across various biological datasets.
Source:
Journal reference:
- Karin, J., Mintz, R., Raveh, B., & Nitzan, M. (2024). Interpreting single-cell and spatial omics data using deep neural network training dynamics. Nature Computational Science, 4(12), 941-954. DOI: 10.1038/s43588-024-00721-5, https://www.nature.com/articles/s43588-024-00721-5