In a paper published in the Journal of Dairy Science, researchers established a method for rapidly classifying milk products by combining matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) analysis with machine learning (ML) techniques.
 Image Credit: isara design/Shutterstock
Image Credit: isara design/Shutterstock
They used two milk product types as examples and applied six feature selection techniques with a support vector machine (SVM) classifier to identify crucial variables and classify the samples. The models were evaluated based on accuracy, akaike information criterion (AIC), and Bayesian information criterion (BIC). The least absolute shrinkage and selection operator (LASSO) combined with SVM showed the highest efficiency. It produced results characterized by favorable AIC and BIC values alongside high accuracy.
Background
Previous studies have extensively explored various techniques for authenticating milk, such as chromatography-mass spectrometry. These approaches are pivotal in ensuring the quality and authenticity of dairy products across diverse contexts and regulatory frameworks. Still, these limitations include slow processing and the need for skilled operators. These methods also require extensive sample preparation and are costly. Additionally, they may not effectively capture all relevant chemical information from mass spectra.
Experimental Setup Overview
Ninety milk samples were sourced from local markets in Nanjing, China, and purchased over two months from nine supermarkets. Ultrapure water from Millipore was used to prepare all aqueous solutions. Each milk sample underwent dilution and mixing with SA matrix solution before being applied to stainless steel target plates for drying.
The analysts analyzed using an applied biosystems sciex (AB Sciex) 4800 Plus MALDI-TOF/TOF mass spectrometer in positive ion linear mode with a 355 nm Nd: YAG laser. The team processed mass spectral data into a structured comma-separated file (CSV) for analysis within a Python environment.
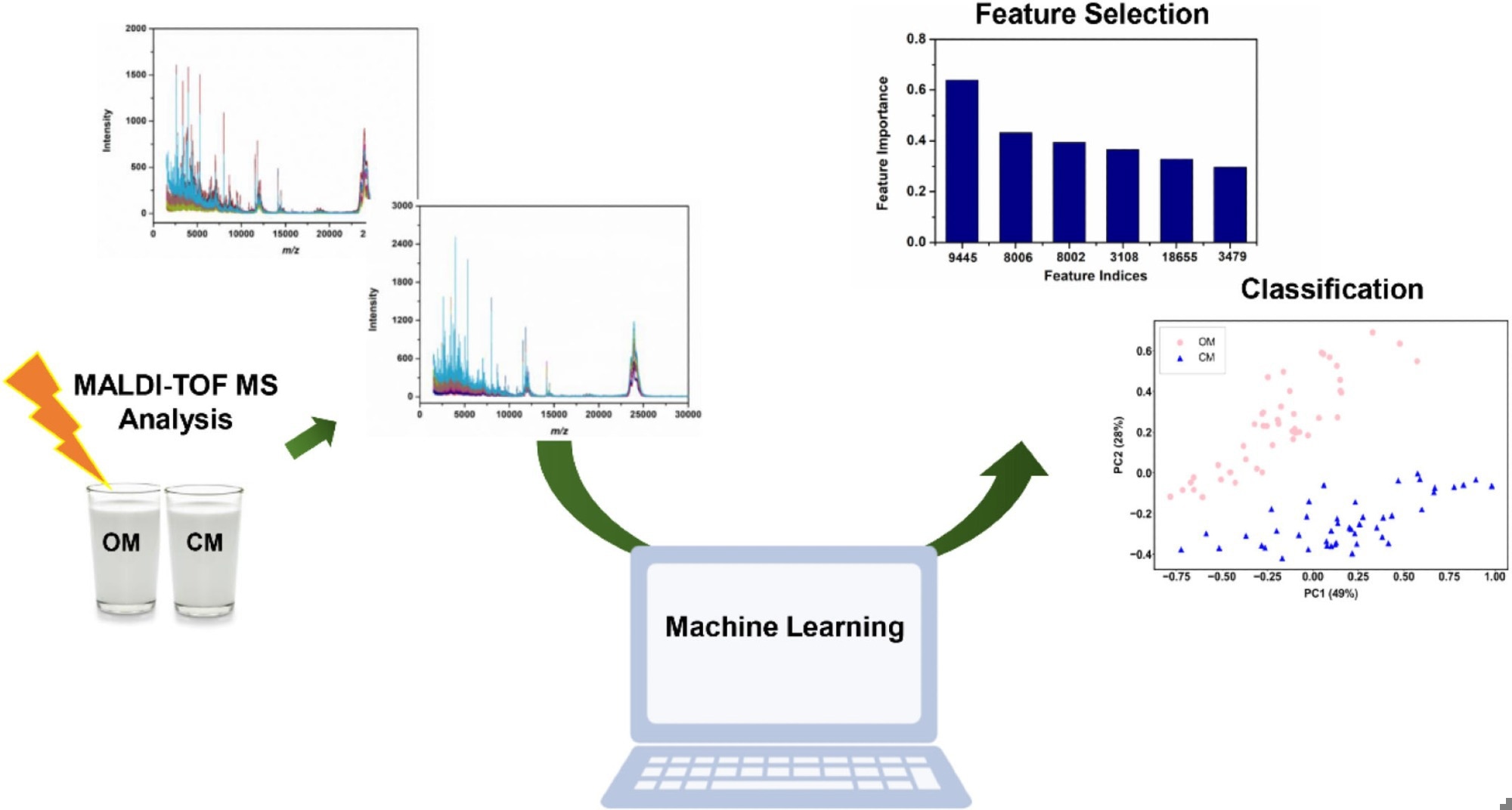 Schematic representation of the proposed machine learning-assisted MALDI-TOF MS method for rapid classification of milk products. Image Credit: https://www.sciencedirect.com/science/article/pii/S0022030224009494
Schematic representation of the proposed machine learning-assisted MALDI-TOF MS method for rapid classification of milk products. Image Credit: https://www.sciencedirect.com/science/article/pii/S0022030224009494
The principal component analysis (PCA) visualized dataset structure, while LASSO, chi-squared, Pearson correlation, information gain, analysis of variance, and elastic net identified discriminatory features. SVM models were trained on selected features to classify milk types, optimizing hyperparameters via grid search cross-validation (GridSearchCV) for accuracy, AIC, and BIC evaluation.
Milk Differentiation Study
This study's objective was to utilize molecular mass data to evaluate the chemical compositions of two types of milk. The goal was to differentiate organic milk (OM) from conventional milk (CM) based on these molecular profiles. Milk samples underwent dilution with ultrapure water, mixing with an SA matrix solution, and analysis using MALDI-TOF MS in positive ion linear mode across a mass range of 1500 Da to 30 kDa.
Representative spectra displayed distinct peaks corresponding to known milk proteins, revealing consistent patterns with minor intensity variations between OM and CM. This approach highlighted the potential of MALDI-TOF MS for identifying milk types based on protein quantification, albeit with complexities in standardization compared to statistical methods.
PCA was used to show the heterogeneity within the MALDI-TOF MS data set. A 2D PCA score plot demonstrated a reasonable separation between OM and CM samples, primarily driven by PC1, which accounted for 77% of the variance. While the plot indicated the potential for distinguishing milk types, some overlap was observed, suggesting nuances in protein profiles that statistical methods could elucidate.
Six different techniques were used to select features from the intricate mass spectrometric data. This method illustrated the efficacy of LASSO in feature selection and was crucial for identifying discriminative mass spectrometric peaks essential for developing SVM models to classify OM and CM samples. These peaks were further analyzed to understand their significance in differentiating milk types, emphasizing their potential as markers influenced by farming practices and environmental factors.
To summarize, integrating MALDI-TOF MS with ML techniques, particularly LASSO for feature selection, effectively characterizes and distinguishes between OM and CM based on their molecular profiles. The study underscores the importance of selected mass spectrometric features in reflecting nuanced differences in protein composition, which are essential for milk authentication and quality assurance in the dairy industry.
Conclusion
In conclusion, the study established a method integrating MALDI-TOF MS profiling and ML to discriminate between milk products rapidly. The LASSO model outperformed other methods in selecting six key features (m/z 9445, 8006, 8002, 3108, 18655, 3479) crucial for distinguishing OM from CM. Statistical analysis revealed differences in mass spectrometry intensity between OM and CM, suggesting these features as potential markers for quality control in the dairy industry.
Subsequent investigations are intended to validate these markers and elucidate their biological significance in the context of milk product differentiation. This approach holds the potential for extending its application to enhance food quality assurance practices across diverse geographical regions and various food product categories. The insights gleaned from this study contribute significantly to developing robust tools for authenticating and ensuring the quality control of food products, thereby addressing critical consumers and industries.