In an article recently published in the journal Nature Communications, researchers proposed a tetrahedral nanostructure-based natronobacterium gregoryi argonaute (NgAgo) on wearable microneedles for long-term ultratrace nucleic acids monitoring.
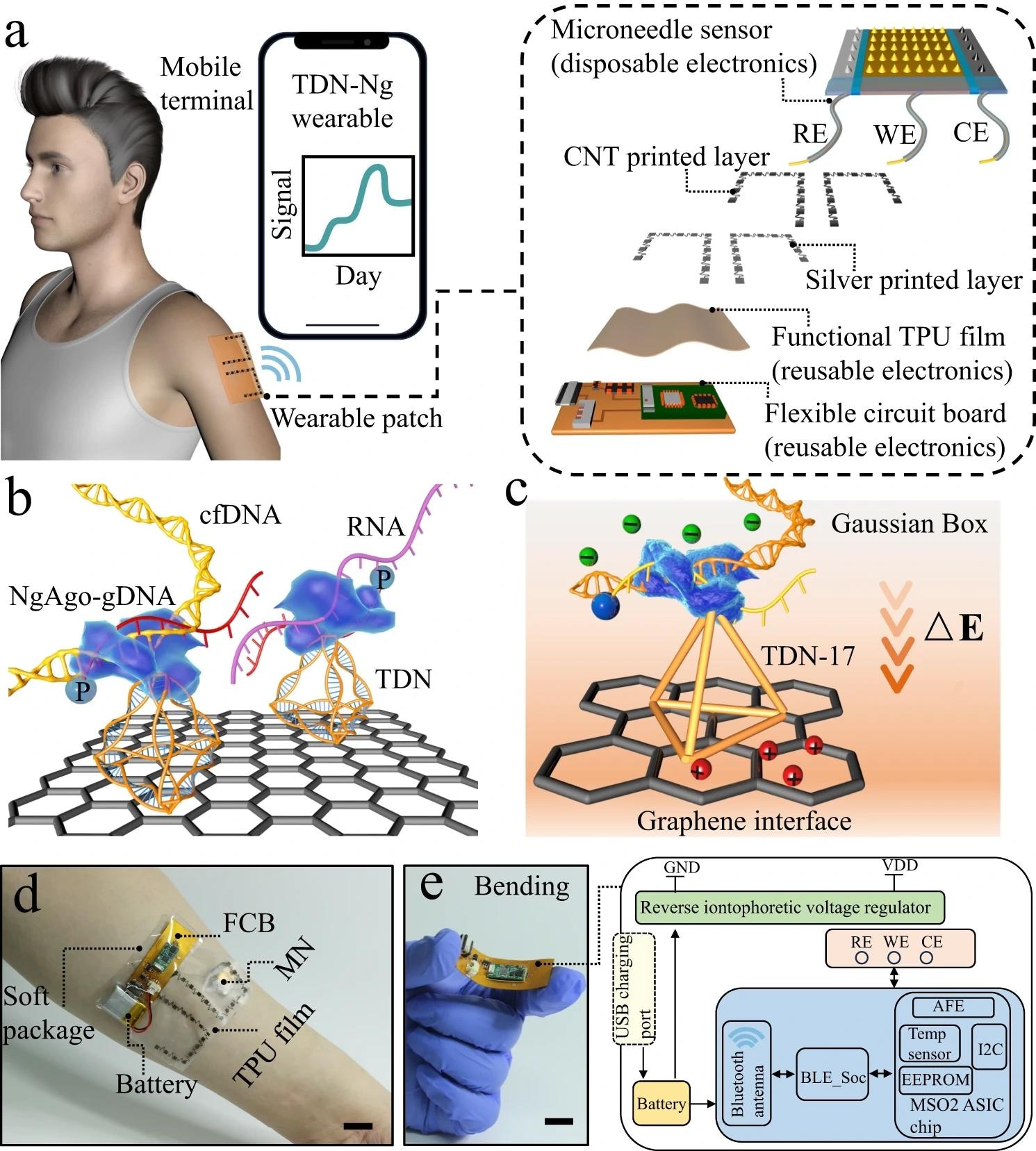 Overview of the integrated wearable system based on TDN-Ng for real-time monitoring of nucleic acids. a) Workflow and view of the wearables on patients’ epidermis for practical use. All the data were transmitted to an electronic device, RE, WE, and CE referring to the reference, working, and counter electrodes, respectively. b) Illustration showing the engineered biosensing interface on a microneedle, in which TDN is used as an anchor to combine with graphene and NgAgo-gDNA, P referring to phosphorylated. c) Schematic of the biosensing mechanism. The enhanced TDN-Ng interfaces selectively recognize target nucleic acids and produce different potentials based on Gaussian box theory, generating electrochemical signal outputs. d) A photograph of the whole integrated wearable system, which was worn on the patient’s arm. Scale bar, 1 cm. e) Block diagram of the flexible circuit board electronics. The electrochemical analog front end (AFE) performs electrochemical sensing to acquire the signal, and the Bluetooth low-energy (BLE) module processes the signal and transmits the data to an outer electronic device via a Bluetooth antenna. The reverse iontophoresis module is individually connected with a battery. The battery was connected to the board and USB charging port. Scale bar, 1 cm. https://www.nature.com/articles/s41467-024-46215-w
Overview of the integrated wearable system based on TDN-Ng for real-time monitoring of nucleic acids. a) Workflow and view of the wearables on patients’ epidermis for practical use. All the data were transmitted to an electronic device, RE, WE, and CE referring to the reference, working, and counter electrodes, respectively. b) Illustration showing the engineered biosensing interface on a microneedle, in which TDN is used as an anchor to combine with graphene and NgAgo-gDNA, P referring to phosphorylated. c) Schematic of the biosensing mechanism. The enhanced TDN-Ng interfaces selectively recognize target nucleic acids and produce different potentials based on Gaussian box theory, generating electrochemical signal outputs. d) A photograph of the whole integrated wearable system, which was worn on the patient’s arm. Scale bar, 1 cm. e) Block diagram of the flexible circuit board electronics. The electrochemical analog front end (AFE) performs electrochemical sensing to acquire the signal, and the Bluetooth low-energy (BLE) module processes the signal and transmits the data to an outer electronic device via a Bluetooth antenna. The reverse iontophoresis module is individually connected with a battery. The battery was connected to the board and USB charging port. Scale bar, 1 cm. https://www.nature.com/articles/s41467-024-46215-w
Background
Genetic testing techniques allow related disease identification through nucleic acid-based diagnostics. Established tools such as whole genome sequencing and polymerase chain reaction (PCR) can provide physiological information that is indicative of several pathologies. However, the need for bulky equipment and long turnover time remain the major limitations of genome sequencing and PCR.
Although several amplification-free methodologies have been recently developed for detecting unamplified target nucleic acids, addressing the limitations of conventional nucleic acid molecular diagnosis, these methodologies require sensitive, hands-free, and continuous monitoring. Fully integrated wearable techniques can bridge this gap.
Wearable technology for monitoring
Wearable technology can ensure simultaneous and continuous monitoring of several biomarkers on the human epidermis, which makes the technology suitable for real-time disease diagnosis, internet-of-medical-things, and personalized medicine. Specifically, state-of-the-art (SOTA) wearable devices have received significant attention as they can monitor the physiological information of subjects, including deep tissue tracking using artificial intelligence (AI) and detecting tumor regression.
Continuous and real-time monitoring of nucleic acid biomarkers using wearable devices holds substantial potential for personal health management, specifically in the context of pandemic surveillance or intensive care unit (ICU) disease, such as monitoring ICU patients with sepsis.
However, achieving long-term stability and high sensitivity is a major challenge in this approach. Specifically, the existing wearable devices cannot effectively perform long-term and real-time monitoring of low-abundance disease-related cell-free deoxyribonucleic acid (cfDNA) or ribonucleic acid (RNA) biomarkers.
The proposed approach
In this study, researchers proposed a fully integrated wearable electronics based on prokaryotic argonaute technology and tetrahedral DNA nanostructures (TDNs) for universal real-time nucleic acid monitoring and sepsis-associated intervention caused by Pseudomonas aeruginosa (PA), Staphylococcus aureus (SA), and EBV. Specifically, a tetrahedral nanostructure-based NgAgo was used for stable long-term monitoring of ultratrace unamplified nucleic acids/cell-free DNAs and RNAs in vivo for sepsis on wearable devices.
The integrated wireless wearable system contained an engineered microneedle biosensor, a stretchable epidermis patch with enrichment capability, and a flexible circuit board. TDN scaffolds were utilized to achieve ultrasensitive detection, while the engineered single-stranded DNA-guided NgAgo biorecognition interface (NgAgo-gDNA) facilitated the system's long-term stability.
The integrated wearable system (TDN-NgAgo-gDNA/TDN-Ng) was used for real-time sepsis-related RNA and cfDNA tracking in interstitial fluid (ISF) to monitor sepsis for ICU onset. Researchers evaluated the TDN-Ng wearable system in vitro to determine its effectiveness in achieving real-time target nucleic acid monitoring in practical scenarios as an integrated wearable electronic device both in vivo and in vitro. They assessed the system's anti-interference properties, specificity, long-term stability, quantitative detection, and sensitivity.
Research findings
The fabricated TDN-Ng microneedles effectively monitored various target cfDNA concentrations in real-time sensitively and quantitatively in vitro. The target cfDNA was recognized continuously and bound by the engineered Ng system on the microneedle surface.
Additionally, the proposed TDN-based NgAgo-gDNA system displayed significantly faster reaction kinetics compared to previously reported CRISPR microneedle patches. Thus, the platform could be used for EBV cfDNA's endpoint quantitative detection. Moreover, the TDN-Ng microneedle platform/patch with the engineered Ng system's biosensing element realized long-term stability.
It demonstrated a long-term stability of 16 days within reasonable coefficient of variation (CV) values in vitro. The TDN-Ng microneedle patch also displayed satisfactory selective target EBV cfDNA detection in interfering with kidney transplantation cfDNA and sepsis cfDNA. It could withstand 80% fetal bovine serum (FBS) and 0.3 nM nonspecific nucleic acid interference.
The TDN-Ng microneedle system effectively distinguished NTC and positive groups and reliably monitored the dynamic changes/fluctuations in target RNAs and cfDNA in vivo in a multiple, accurate, quantitative manner. It displayed similar reliability as the PCR method while recording the dynamic changes in target cfDNA in vivo.
Additionally, the TDN-Ng microneedle displayed stability over 14 days with 4.87% C.V., which is longer than the median ICU stay duration of 11 days for sepsis patients. Thus, the 14-day stability of the TDN-Ng microneedle device for real-time nucleic acid monitoring in vivo met the clinical requirements for ICU patients.
Moreover, the TDN-Ng patch with specific gDNA successfully monitored two kinds of sepsis mice bearing SA and PA within the inflammatory cytokine storm period. In vivo experiments also demonstrated the suitability for real-time monitoring of cell-free DNA and RNA with a sensitivity of 0.3 fM for up to 14 days. To summarize, the findings of this study demonstrated the effectiveness of the proposed integrated, wireless wearable electronics for highly sensitive molecular recognition in vivo and for on-body detection of nucleic acid.
Journal reference:
- Yang, B., Wang, H., Kong, J., Fang, X. (2024). Long-term monitoring of ultratrace nucleic acids using tetrahedral nanostructure-based NgAgo on wearable microneedles. Nature Communications, 15(1), 1-15. https://doi.org/10.1038/s41467-024-46215-w, https://www.nature.com/articles/s41467-024-46215-w