Scientists have revolutionized microscopy imaging with μSAM, a cutting-edge AI tool that surpasses existing methods, making complex biological image segmentation faster, more precise, and accessible to researchers worldwide.
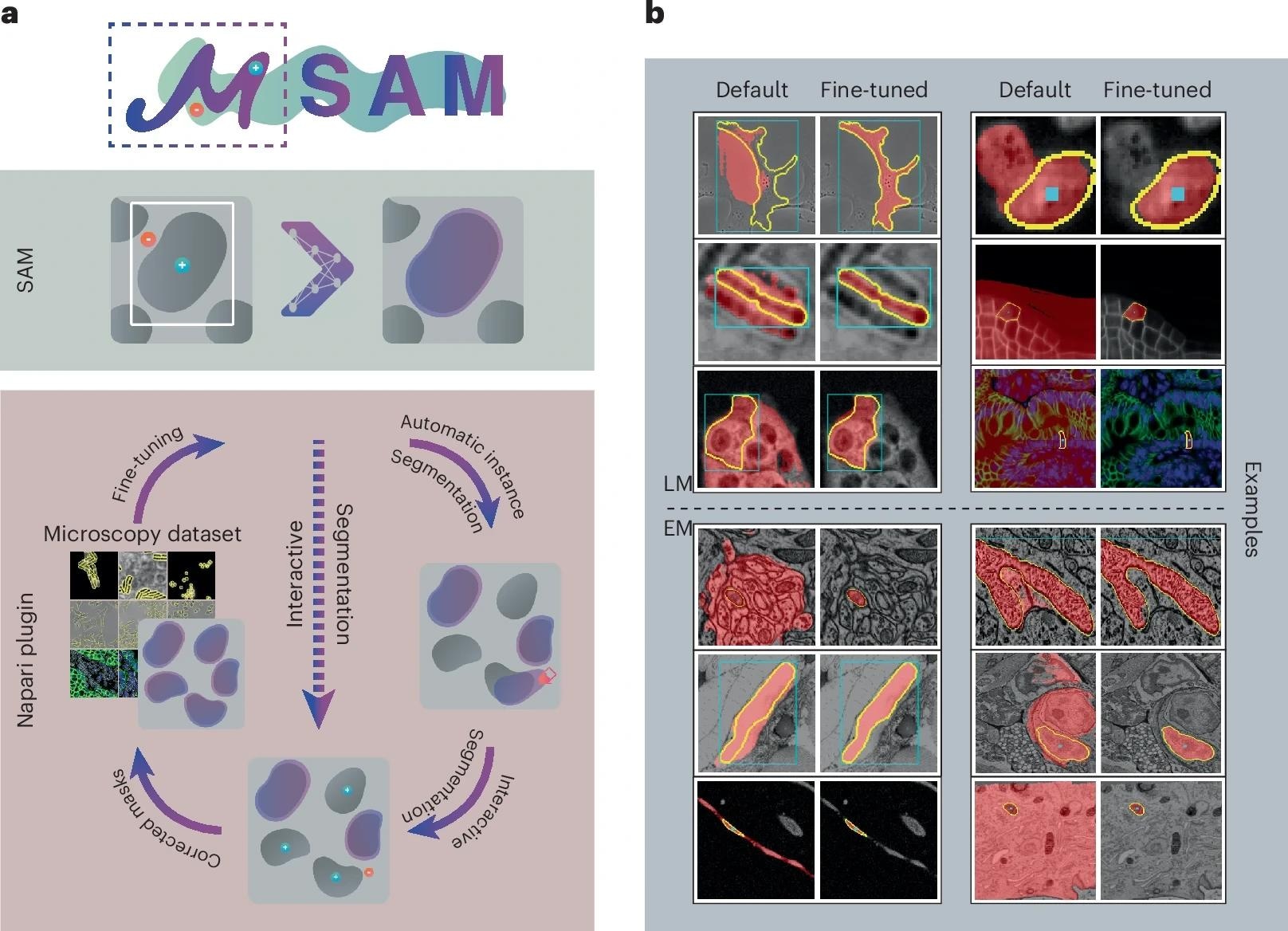 a, We provide a napari plugin for segmenting multidimensional microscopy data. This tool uses SAM, including our improved models for LM and EM (see b). It supports automatic and interactive segmentation as well as model retraining on user data. The drawing sketches a complete workflow based on automatic segmentation, correction of the segmentation masks through interactive segmentation and model retraining based on the obtained annotations. Individual parts of this workflow can also be used on their own, for example, only interactive segmentation can be used as indicated by the dashed line. b, Improvement of segmentation quality due to our improved models for LM (top) and EM (bottom). Blue boxes or blue points show the user input, yellow outlines show the true object and red overlay depicts the model prediction.
a, We provide a napari plugin for segmenting multidimensional microscopy data. This tool uses SAM, including our improved models for LM and EM (see b). It supports automatic and interactive segmentation as well as model retraining on user data. The drawing sketches a complete workflow based on automatic segmentation, correction of the segmentation masks through interactive segmentation and model retraining based on the obtained annotations. Individual parts of this workflow can also be used on their own, for example, only interactive segmentation can be used as indicated by the dashed line. b, Improvement of segmentation quality due to our improved models for LM (top) and EM (bottom). Blue boxes or blue points show the user input, yellow outlines show the true object and red overlay depicts the model prediction.
Identifying and delineating cell structures in microscopy images is crucial for understanding the complex processes of life. This task is called "segmentation," and it enables both automatic and interactive image analysis, allowing researchers to refine and improve results with minimal manual effort. It enables a range of applications, such as analyzing the reaction of cells to drug treatments or comparing cell structures in different genotypes. It was already possible to carry out automatic segmentation of those biological structures, but the dedicated methods only worked in specific conditions, and adapting them to new conditions was costly.
An international research team led by Göttingen University has now developed a technique by fine-tuning the existing AI-based software Segment Anything with a new decoder and iterative training scheme on over 17,000 microscopy images with over 2 million structures annotated by hand. Their new model is called Segment Anything for Microscopy (μSAM), which significantly enhances segmentation accuracy and versatility across light and electron microscopy. It can precisely segment images of tissues, cells, and similar structures in a wide range of settings. To make it available to researchers and medical doctors, they have also created μSAM, a user-friendly software tool integrated into napari, a popular microscopy image viewer. This tool enables both automatic and interactive segmentation, allowing users to refine segmentation with minimal manual input. Their work was published in the journal Nature Methods.
To adapt the existing software to microscopy, the research team first evaluated it on a large set of open-source data, which showed the model's potential for microscopy segmentation. To improve quality, the team implemented an advanced fine-tuning approach, retraining it on a large microscopy dataset. This dramatically improved the model's performance for the segmentation of cells, nuclei, and tiny structures in cells known as organelles. Unlike the original SAM, which struggled with distinguishing clustered cells, μSAM introduces a more precise segmentation approach by integrating improved instance segmentation techniques. The team then created their software, μSAM, which enables researchers and medical doctors to analyze images without the need to first manually paint structures or train a specific AI model. The software is already widely used internationally, for example, to analyze nerve cells in the ear as part of a project on hearing restoration, to segment artificial tumor cells for cancer research, or to analyze electron microscopy images of volcanic rocks.
"Analysing cells or other structures is one of the most challenging tasks for researchers working in microscopy and is an important task for both basic research in biology and medical diagnostics," says Junior Professor Constantin Pape at Göttingen University's Institute of Computer Science. "My group specializes in building tools to automate such tasks and we often get asked by researchers to help. Before the development of Segment Anything for Microscopy, we had to ask them to first annotate a lot of structures by hand – a difficult and time-consuming task. μSAM has changed this! Tasks that used to take weeks of painstaking manual effort can be automated in a few hours, because the model can segment any kind of biological structure with a few clicks and can then be further improved to automate the task with our tool."
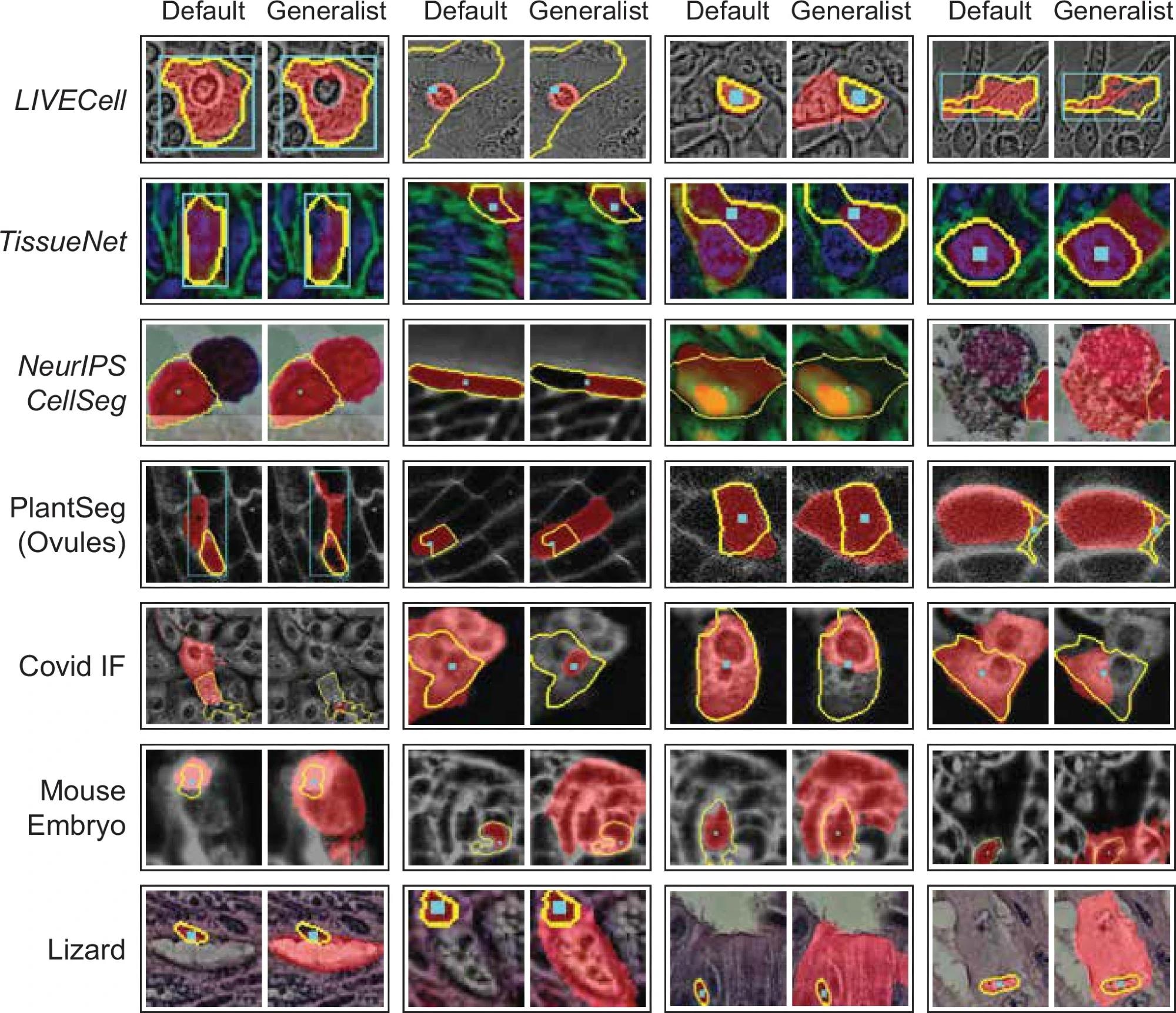 Qualitative comparison of interactive segmentation for the default SAM and our LM generalist (ViT-L). In the opposite approach, we show the objects where the decrease in IOU is largest, comparing the generalist and default models. Here, we see a few different effects: in some cases the generalist model segments several nearby cells (proving an exception to the general behavior observed previously) for point annotations, in other cases the segmentation quality is lower because the generalist segments smaller sub-structures. This systematic effect can also be observed for Covid IF, where the generalist often segments only the nucleus, which is discernible from the rest of the cell, rather than the full cell. Note that the quantitative segmentation quality for all these datasets is clearly higher for the generalist model.
Qualitative comparison of interactive segmentation for the default SAM and our LM generalist (ViT-L). In the opposite approach, we show the objects where the decrease in IOU is largest, comparing the generalist and default models. Here, we see a few different effects: in some cases the generalist model segments several nearby cells (proving an exception to the general behavior observed previously) for point annotations, in other cases the segmentation quality is lower because the generalist segments smaller sub-structures. This systematic effect can also be observed for Covid IF, where the generalist often segments only the nucleus, which is discernible from the rest of the cell, rather than the full cell. Note that the quantitative segmentation quality for all these datasets is clearly higher for the generalist model.
"In comparative evaluations, μSAM demonstrated superior segmentation accuracy over widely used tools such as CellPose, ilastik, and TrackMate, particularly in segmenting cell nuclei and organelles. This competitive performance highlights its potential as a leading tool for microscopy image analysis."
The research team acknowledges that μSAM is not without limitations. While it provides a major leap forward in general segmentation capabilities, developing a truly universal model that works equally well across all microscopy domains remains a challenge. The team plans to further refine the model to enhance its performance in electron microscopy and expand its training datasets for even broader applicability.
"This enables many new applications," Pape continues, "and we have already used it in a wide range of projects, ranging from basic cell biology to developing tools for treatment recommendation in cancer therapies. With continued improvements, we hope μSAM will become an essential tool for researchers working across various imaging modalities."
Source:
Journal reference:
- Archit, A., Freckmann, L., Nair, S., Khalid, N., Hilt, P., Rajashekar, V., Freitag, M., Teuber, C., Buckley, G., Von Haaren, S., Gupta, S., Dengel, A., Ahmed, S., & Pape, C. (2025). Segment Anything for Microscopy. Nature Methods, 1-13. DOI: 10.1038/s41592-024-02580-4, https://www.nature.com/articles/s41592-024-02580-4