In a paper published in the journal Remote Sensing, researchers used deep learning (DL) to segment canola plants from other species in the field, treating all non-canola plants as weeds.
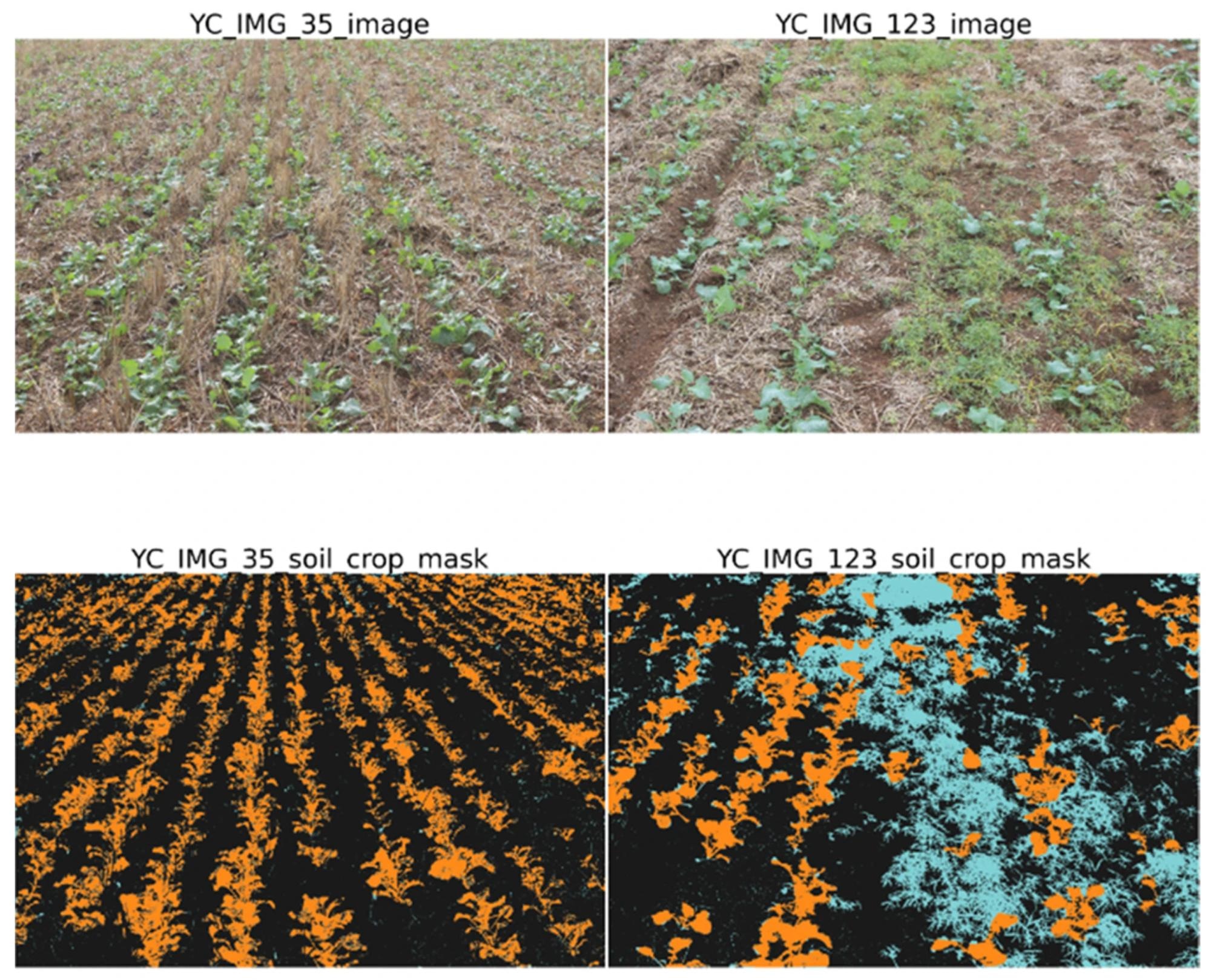 Display of base full size RGB image and labelled crop mask after soil deletion, vegetation index application, and overlay of ground truth labels. Plants other than canola are shown in blue. Image Credit: https://www.mdpi.com/2072-4292/16/11/2041
Display of base full size RGB image and labelled crop mask after soil deletion, vegetation index application, and overlay of ground truth labels. Plants other than canola are shown in blue. Image Credit: https://www.mdpi.com/2072-4292/16/11/2041
They trained models, including residual networks (ResNet-18, ResNet-34) and visual geometry group network (VGG-16), on three datasets (T1_miling, T2_miling, and yellow canola (YC)) containing 3799 images of canola. The best model, ResNet-34, demonstrated superior performance. This approach, which did not require separate weed datasets, highlighted the importance of plant growth stages and positioning for effective weed detection, offering a promising strategy for precision agriculture and innovative weed control.
Related Work
Past work has highlighted significant canola production challenges due to weeds, pests, and diseases, leading to substantial yield losses. Weeds cost growers Australian dollars (AUD 3.3) billion annually in Australia. Herbicide-resistant canola varieties have increased herbicide resistance in ryegrass and wild radish species.
Precision agriculture offers solutions through optimized resource usage and targeted weed management. DL models, especially convolutional neural networks (CNNs) like VGG-16 and ResNet-50, have shown promise in weed identification but face challenges in field conditions. Robust models are needed for effective weed detection and management in commercial farming.
DL for Agriculture
This study utilized three datasets—T1_miling, T2_miling, and YC—comprising images of canola plants in diverse field conditions alongside various other plant species. These datasets were collected using handheld devices and a digital single-lens reflex (DSLR) camera from locations in Western Australia. The images were annotated for ground truth labeling, focusing on delineating canola plants from other vegetation using bounding boxes.
Subsequently, the analysts generated segmentation masks by applying image processing techniques like the color index of vegetation (CIVE) and Otsu Thresholding to differentiate plant and non-plant pixels. Training of DL models—ResNet-18, ResNet-34, and VGG-16—was conducted along with the data augmentation techniques such as flipping, rotation, and zooming to enhance model robustness. The models were evaluated using metrics like the Dice coefficient, Jaccard coefficient, Precision, Recall, and macro F1 score on a holdout set, demonstrating their ability to segment canola plants in complex field conditions accurately.
The training process involved using dual NVIDIA Tesla V100 16 GB graphics processing units (GPUs) for 24 hours per model. Model performance was monitored using callbacks for early stopping and saving the best-performing model weights. Each model was trained using a 5-fold cross-validation strategy to ensure generalizability and robustness across different subsets of the dataset. The team performed feature extraction analysis to understand how ResNet-18, ResNet-34, and VGG-16 architectures encoded features at various layers, providing insights into their learning capabilities.
This study showcases the effectiveness of DL models in accurately segmenting canola plants from diverse backgrounds in field settings. The results highlight the potential of these models for enhancing precision agriculture practices by enabling targeted weed management and resource-efficient crop monitoring.
Segmentation Performance Analysis
The segmentation performance of DL in this study peaked between 30 and 40 epochs, with training sessions lasting approximately 18 hours per model. The models demonstrated consistent prediction capabilities across the T1 Miling, T2 Miling, and YC datasets. Notably, the YC dataset exhibited superior performance metrics, including the highest recall (0.87), intersection over union (IoU) (0.76), and Macro F1 (0.84), attributed to favorable lighting conditions and higher image resolution.
ResNet-34 emerged as the top-performing architecture, achieving an average precision of 0.84, recall of 0.87, IoU of 0.77, and Macro F1 of 0.85. ResNet-18 is closely followed, with slightly lower metrics compared to ResNet-34. In contrast, VGG-16 displayed inferior performance metrics across all datasets, with a precision of 0.80, recall of 0.82, IoU of 0.71, and macro F1 of 0.80. These results indicate that ResNet architectures generally outperformed VGG-16 regarding segmentation accuracy and robustness, with VGG-16 facing constraints such as smaller batch sizes and image resolution modifications.
The feature analysis provided insights into how ResNet-34 and VGG-16 encode features at different layers. For ResNet-34, feature maps at layer 1 showed distinctive patterns related to canola plant structures. In comparison, VGG-16 exhibited feature maps that reflected attention to specific shapes and structures along leaf edges and stems despite its different architectural approach without an encoder/decoder system.
The study underscores the effectiveness of ResNet architectures in accurately segmenting canola plants under varied field conditions. The findings contribute valuable insights into model selection and optimization for precision agriculture applications, emphasizing the importance of architecture choice and feature analysis in enhancing model performance and interpretability.
Conclusion
To summarize, this study used DL techniques to segment canola plants from various field conditions alongside other plant species assumed to be weeds. Using ResNet-18, ResNet-34, and VGG-16 architectures, the best-performing model, ResNet-34, achieved an average precision of 0.84, recall of 0.87, IoU of 0.77, and macro F1 score of 0.85.
The research demonstrated the effectiveness of these models in accurately identifying canola plants, offering a promising approach for weed detection in agricultural settings without requiring separate weed-specific datasets. The findings contribute to advancing precision agriculture methods and suggest potential innovations in weed management practices.