In a paper published in the journal Scientific Reports, researchers analyzed the Cambridge structural database (CSD) to understand lanthanide coordination chemistry, aiming to improve ligand design for rare-earth element (REE) separations. They examined four subsets of lanthanide complexes: all Ln-containing complexes, mononuclear Ln complexes, mononuclear Ln complexes without cyclopentadienyl ligands, and Ln complexes with at least one 1,10-phenanthroline (phen) or derivative.
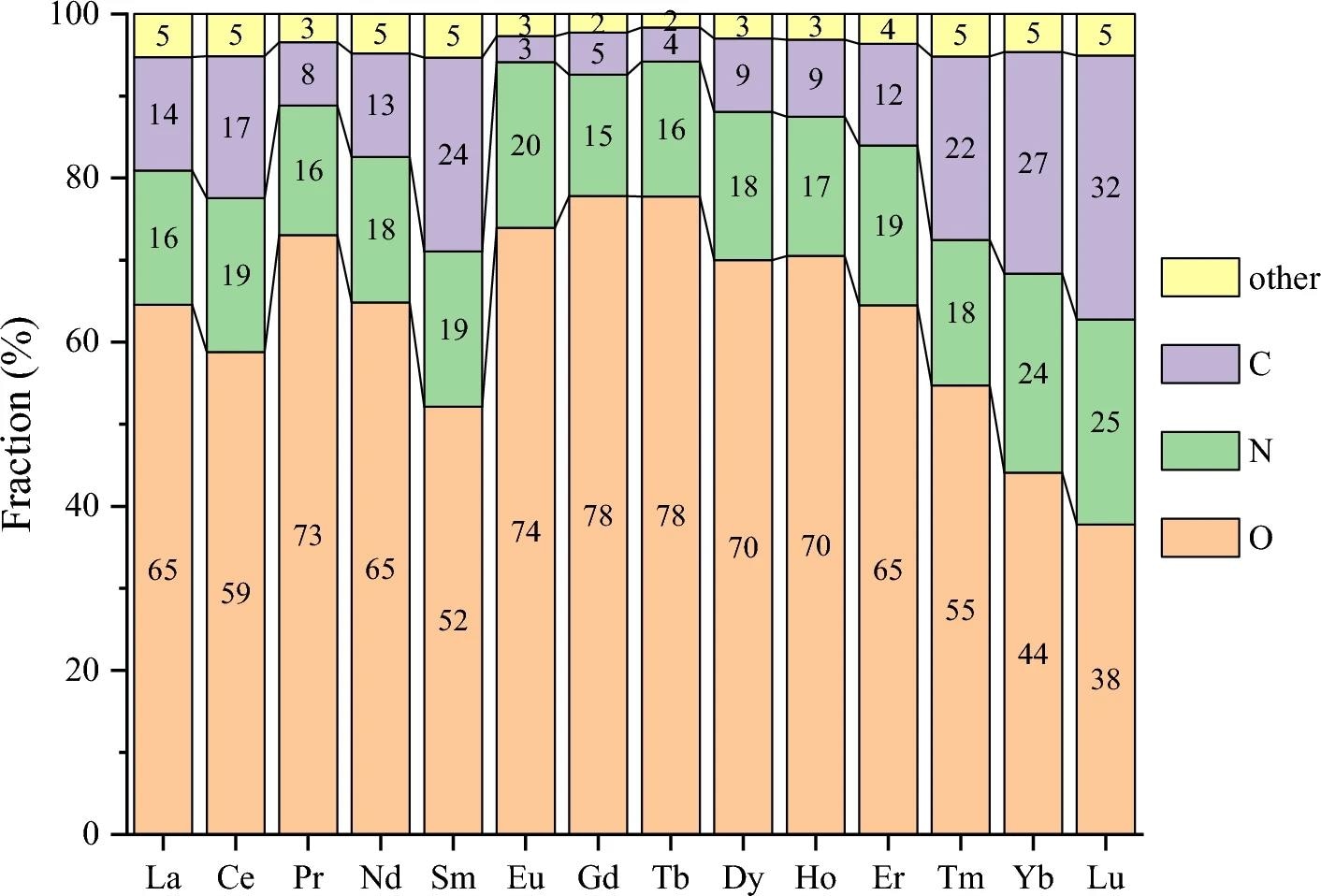 Breakdown of donor types in the first coordination shell for each Ln element in Subset 2. Other elements include H, P, S, F, Cl, Br, and I. Image Credit: https://www.nature.com/articles/s41598-024-62074-3
Breakdown of donor types in the first coordination shell for each Ln element in Subset 2. Other elements include H, P, S, F, Cl, Br, and I. Image Credit: https://www.nature.com/articles/s41598-024-62074-3
The study focused on trends in coordination numbers, first shell distances, and ligand/donor group characterization. Insights from this analysis are intended to inform future data-driven designs of ligands for more efficient REE separation.
Related Work
Past work on REE separations has emphasized their importance in various technologies despite the challenges in separating them due to their similar chemical properties. Solvent extraction has been the primary method for large-scale REE separations since the 1960s. The increasing demand for REEs, particularly electric vehicle motors, has driven research into novel ligand designs, spectroscopic techniques, redox/photo-stimulated processes, ionic liquids, and biomolecule ligands.
With its extensive collection of lanthanide complexes, the CSD offers valuable insights for developing new ligands using machine learning (ML) and artificial intelligence (AI). Inspired by advancements like AlphaFold, researchers are leveraging the CSD to design ligands that enhance REE separations, demonstrating the potential of automated structure creation in this field.
Script-Based Analysis
Using the CSD Python-based applications programming interface (API), the analysts developed task-specific scripts to search and analyze downloaded CSD structures. These scripts retrieved three-dimensional structural information and extracted chemical data from the CSD. The first script focused on capturing the first coordination shell from the original structure to investigate lanthanide coordination numbers.
The second script analyzed the distribution of elements or donor groups within this shell, generating a matrix to record the occurrences of different atoms. The third script identified ligand types binding to the metal center, distinguishing between organic ligands (possessing carbon chains) and inorganic ligands (small molecules like water, nitrate, chloride, and polyoxometalates).
A fourth script specifically targeted lanthanide complexes with phenanthroline and its derivatives as ligands. This module identified complexes with phenanthroline ligands while the team created additional scripts to recognize other specific ligands. The researchers conducted further manual analysis on ligand types and phenanthroline derivatives, counting each cyclopentadienyl ligand as contributing three to the coordination number.
Lanthanide Complex Analysis
After eliminating erroneous entries and those lacking three-dimensional structural information, a total of crystal structures of lanthanide (Ln) complexes in the CSD were identified. A subset of this, comprising mononuclear Ln complex structures, was further analyzed. Notably, the number of structures for praseodymium (Pr), holmium (Ho), thulium (Tm), and lutetium (Lu) is smaller than for other Ln ions.
The average coordination number (CN) decreases across the lanthanide series. The average first shell distance also decreases, reflecting the lanthanide contraction. The exclusion of Cp ligands resulted in a significant decrease in deviations for both CN and first shell distance. Detailed CN distributions for each Ln ion are presented.
The distribution of donor element types in the first coordination shell reveals oxygen atoms as the most common donor groups, followed by carbon and nitrogen atoms. Organic oxygen donors, alkoxide, ether, carboxylate, ketone, and amide ligands are significant contributors. Monodentate ligands are predominant, followed by bidentate ligands.
Commercial complexants analysis identified phosphoric acid ligands as the majority, followed by versatic acids. The distribution across the lanthanide series shows varying numbers of structures for different elements. Analysis of phenanthroline (phen) and its derivatives revealed phen as the most common ligand, with a majority having a 1:1 ligand-to-metal ratio.
While assuming the structures in the datasets are generally accurate, some inaccuracies in the CSD could introduce noise into the data trends. Confirming the accuracy of these structures using quantum chemical methods like density functional theory will be essential. Future efforts should focus on high-throughput automatic structure generation and geometry optimization to build a high-quality structural database for Ln complexes.
Conclusion
To sum up, analyzing lanthanide complexes in the CSD provided valuable insights into their coordination chemistry, donor types, and ligand preferences. A consistent decrease in the average coordination number and first shell distance across the lanthanide series was observed, reflecting the lanthanide contraction phenomenon.
Oxygen donors were predominant in the first coordination shell, followed by carbon and nitrogen. Additionally, the examination of commercial complexants revealed phosphoric acid ligands as the most common choice, with phenanthroline emerging as a prevalent ligand in the studied complexes, often maintaining a 1:1 ratio with the lanthanide ions.
These findings shed light on the structural characteristics of lanthanide complexes and were promising to inform future ligand design approaches for rare-earth separations. By leveraging these structural insights, researchers could pursue data-driven strategies to develop novel ligands tailored for efficient separation of lanthanides, addressing critical needs in various technological applications.