In a paper published in the journal Cell Research, experimenters unveiled new insights into the workings of the suprachiasmatic nucleus (SCN), the central circadian pacemaker in mammals, demonstrating that collective calcium (Ca2+) signals at the population level within the SCN correlated strongly with the time of day.
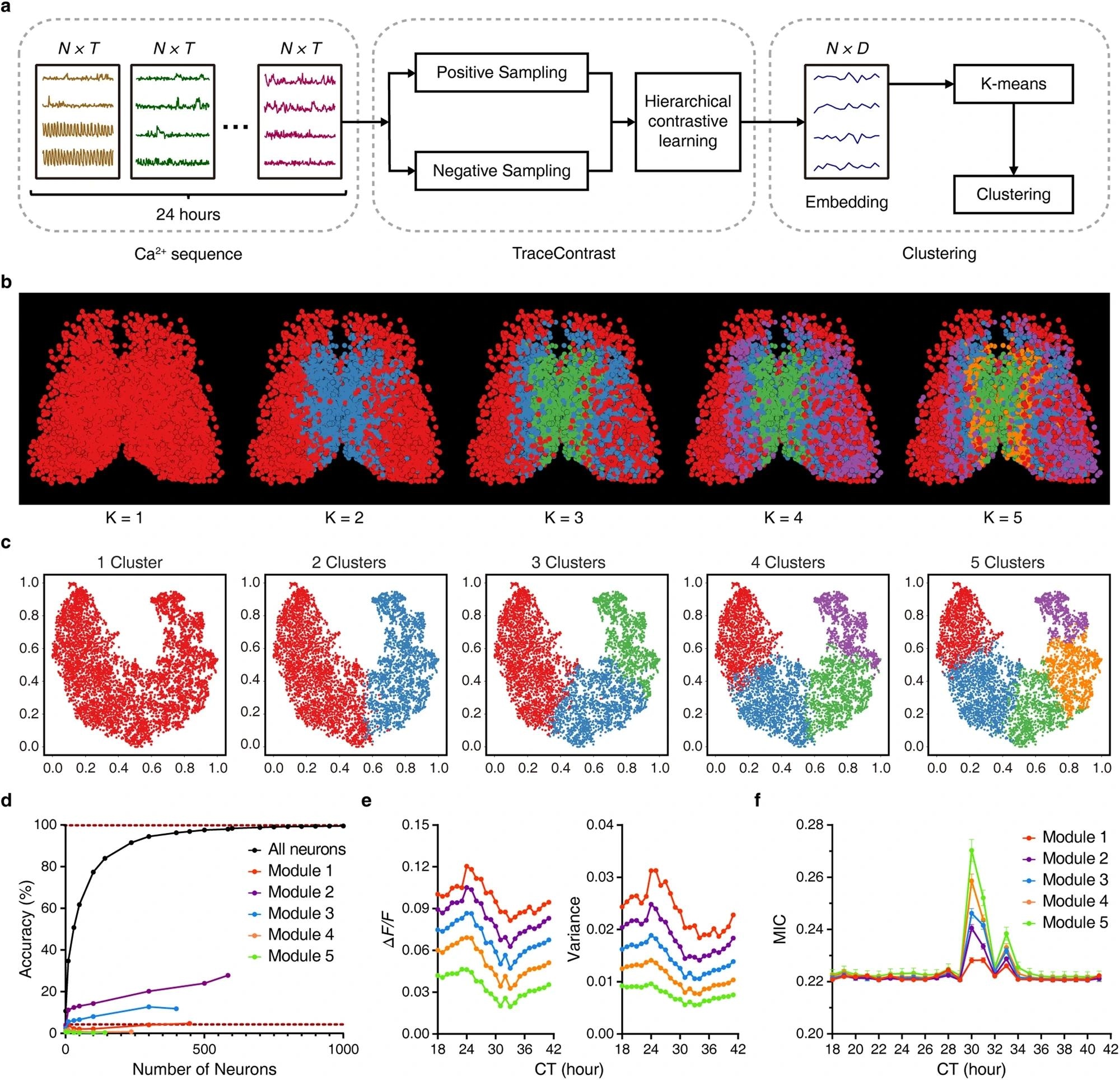 a Scheme of neuron subtype classification by TraceContrast. N, T, and D denote the dimensions of neurons, the input Ca2+ sequence of each neuron and its output features, respectively. b Bilaterally symmetric and hierarchical modularity emerged from neuron subtype classification. The predefined number of clusters (K) is listed below each corresponding image. c The t-SNE plots of the dimensionality reduction corresponding to b. d Time predictability when sampling only within one specific module (K = 5). Lower dashed line represents the chance level. Data are shown as mean ± SEM (n = 5000 trials). Note that one-module-only sampling results in a marked disruption to time prediction accuracy compared to random sampling in all SCN neurons. Similar results were obtained when K = 2, 3, 4. e, f Ca2+ signal amplitude and corresponding variance (e) and average MIC in different modules (f). Data are shown as mean ± SEM (n = 1587, 1936, 1211, 835, and 480 for Modules 1, 2, 3, 4 and 5, respectively). Note that the two attributes showed smooth inter-modular gradients running in opposite directions. Image Credit: https://www.nature.com/articles/s41422-024-00956-x
a Scheme of neuron subtype classification by TraceContrast. N, T, and D denote the dimensions of neurons, the input Ca2+ sequence of each neuron and its output features, respectively. b Bilaterally symmetric and hierarchical modularity emerged from neuron subtype classification. The predefined number of clusters (K) is listed below each corresponding image. c The t-SNE plots of the dimensionality reduction corresponding to b. d Time predictability when sampling only within one specific module (K = 5). Lower dashed line represents the chance level. Data are shown as mean ± SEM (n = 5000 trials). Note that one-module-only sampling results in a marked disruption to time prediction accuracy compared to random sampling in all SCN neurons. Similar results were obtained when K = 2, 3, 4. e, f Ca2+ signal amplitude and corresponding variance (e) and average MIC in different modules (f). Data are shown as mean ± SEM (n = 1587, 1936, 1211, 835, and 480 for Modules 1, 2, 3, 4 and 5, respectively). Note that the two attributes showed smooth inter-modular gradients running in opposite directions. Image Credit: https://www.nature.com/articles/s41422-024-00956-x
Employing advanced imaging techniques and machine learning (ML) algorithms, groups of SCN neurons collectively achieved accurate hourly time predictions. Additionally, the study revealed distinct functional subtypes of neurons within the SCN, which tended to cluster together in spatially organized modules, creating ripple-like patterns. These modules represented specific time features, enabling accurate time decoding even within individual modules, shedding new light on the intricate mechanisms underlying the biological clock's design principles at a systemic level.
Background
Past work on the SCN, the central pacemaker of the mammalian circadian rhythm, has highlighted its complexity and importance in regulating physiological functions and daily behaviors. However, scientists have yet to discover the exact mechanism by which the SCN computes and represents time. SCN neurons are heterogeneous and exhibit spatiotemporal gradients in clock gene expression and intracellular signaling.
Despite extensive research, fundamental questions persist regarding how to decode these gradients and specificities into the time of day and whether all SCN neurons contribute uniformly to this process. Moreover, the technology and methodology needed to decipher the design principles of complex nuclei like the SCN have been lacking.
Experimental Procedure Overview
The study involved generating and selecting mice expressing green calcium-modulated protein 6s (GCaMP6s) or GCaMP6f, specifically in gamma-aminobutyric acid (GABAergic) neurons, for subsequent experiments, adhering to animal care guidelines.
SCN slices were prepared from male mice aged 4-6 weeks individually housed with running wheels to assess locomotor activity. After perfusion and brain removal, 300-μm coronal slices containing the middle-rostrocaudal region of the SCN were prepared and incubated for subsequent Ca2+ imaging.
The analysts conducted Ca2+ imaging of SCN slices using two-photon microscopy, with fluorescence signals reported by GCaMP6f. A custom-built dual-view two-photon microscope achieved volumetric Ca2+ imaging of the entire 300-μm SCN slice. The resulting data underwent a 3D-t image analysis pipeline involving motion correction, image registration, region of interest (ROI) segmentation, and neuronal Ca2+ time series extraction.
Graph-based classification was applied to classify Ca2+ states of SCN neurons, followed by visualization techniques to depict spatiotemporal patterns of Ca2+ signals. Researchers developed a time predictor based on Ca2+ signals using a convolutional neural network (CNN) and quantified the contribution coefficient of each neuron to time prediction.
Functional neuron subtypes were classified using hierarchical contrasting, followed by visualizing their modular organization in 3D. The investigation team analyzed the experimental data statistically, assessing significance using appropriate methods such as the Wilcoxon rank sum test.
Intricate SCN Dynamics
The analysis delved into the intricate dynamics of Ca2+ signals within the SCN, the central pacemaker of the mammalian circadian system, by examining SCN slices from adult mice expressing the Ca2+ indicator GCaMP6f in GABAergic neurons through two-photon microscopy. Diverse Ca2+ bursts, varying in frequency, amplitude, duration, and waveform, were observed among these neurons. Clustering algorithms categorized these bursts into five distinct classes, while six Ca2+ states exhibited circadian rhythmicity across the neuronal population.
Advanced imaging techniques, including a dual-view two-photon microscope, were developed to extend observations and capture Ca2+ dynamics over prolonged periods. The team leveraged ML to train a CNN for hourly time prediction based on Ca2+ signals from randomly selected cohorts of neurons. Time prediction accuracy increased with the neuronal cohorts' size, highlighting the collective nature of time representation in the SCN.
Further analysis revealed a modular organization of functional neuron subtypes within the SCN. Using contrastive learning methods, researchers classified neurons into distinct subtypes, each forming ripple-like modules with bilateral symmetry. These modules exhibited specific patterns of Ca2+ signal amplitude and coupling strength, indicating unique representations of time features. Training time predictors with data from particular modules resulted in accurate same-module time prediction but impaired cross-module prediction, emphasizing the topological specificity of time feature representation.
The modular organization of functional subtypes was robust against factors such as PWHA and spatial/temporal subsampling. This organization provides diverse and finely tuned time representation within the SCN, potentially facilitating differential outputs to downstream brain areas and peripheral clocks. Additionally, the study highlighted the dynamic load-balancing mechanism among SCN neurons, ensuring uniform contribution to time computation over 24 hours.
The study unveiled the intricate spatiotemporal dynamics of Ca2+ signals in the SCN, demonstrating its role in encoding and computing time information. The team gained insights into the collective nature and modular organization of time representation in the SCN through advanced imaging techniques and ML approaches. These findings deepen the understanding of the mammalian circadian time-keeping system and have implications for diverse physiological processes regulated by the circadian clock.
Conclusion
In conclusion, this study advanced the understanding of the mammalian circadian system by revealing the diverse Ca2+ dynamics within the SCN. The investigation elucidated the collective nature of time representation in the SCN through innovative imaging techniques and ML approaches, emphasizing its modular organization and topological specificity. These findings had implications for various physiological processes regulated by the circadian clock.