In a recent article published in the journal Nature Communications, researchers from China introduced and validated an innovative method called protein language model search (PLMSearch) for enhancing sensitivity and accuracy in detecting remote homologous proteins.
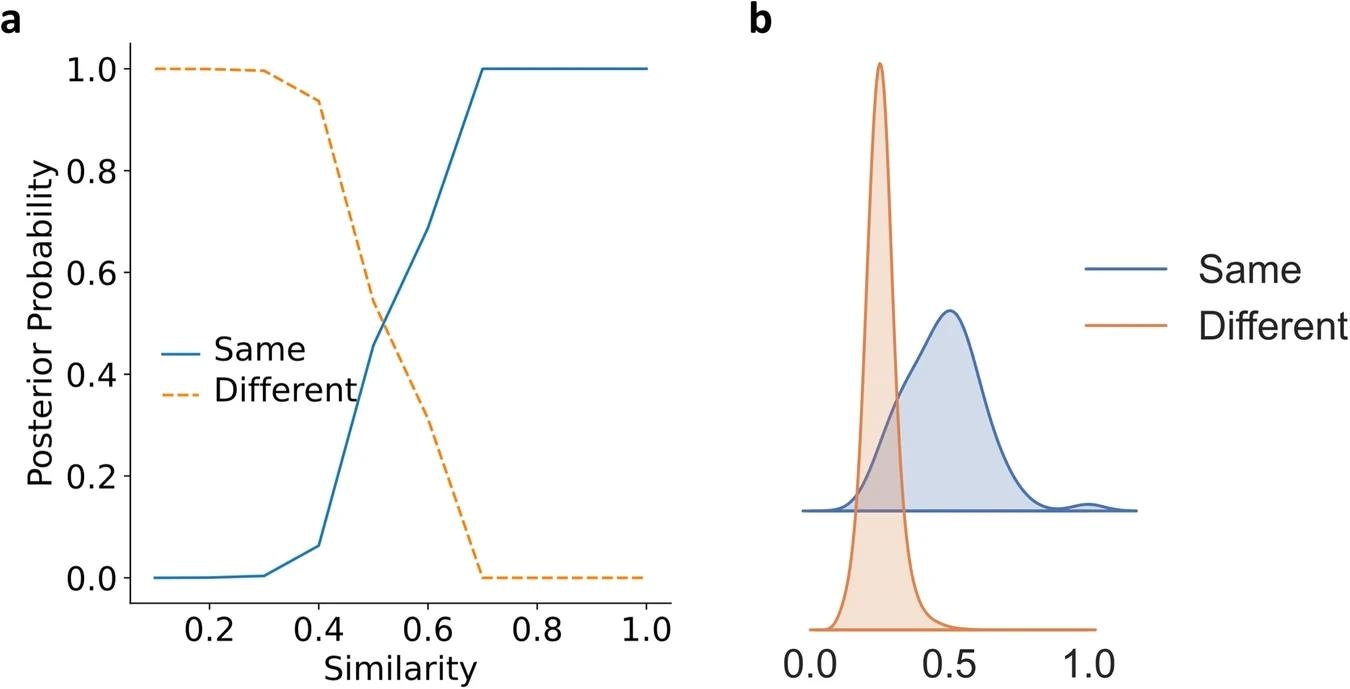 a The posterior probability of proteins with a given similarity being in the same fold or different folds in SCOPe40-train. b Similarity distribution of the same and different folds protein pairs using kernel density estimation (smoothed histogram using a Gaussian kernel with the width automatically determined). Image Credit: https://www.nature.com/articles/s41467-024-46808-5
a The posterior probability of proteins with a given similarity being in the same fold or different folds in SCOPe40-train. b Similarity distribution of the same and different folds protein pairs using kernel density estimation (smoothed histogram using a Gaussian kernel with the width automatically determined). Image Credit: https://www.nature.com/articles/s41467-024-46808-5
They comprehensively addressed the limitations of traditional methods by leveraging deep representations from a pre-trained protein language model to improve remote homology detection, particularly focusing on identifying evolutionary relationships from sequences that exhibit significant structural similarities despite sequence divergence.
Background
Protein homology search is a fundamental aspect of bioinformatics crucial for understanding protein function, structure, and evolution. Traditional methods often rely on structural information, which can be limited in detecting distant relationships solely from sequences. It struggles to identify distant evolutionary relationships solely based on sequence information.
The advent of deep learning models has revolutionized this field by leveraging advanced algorithms to enhance the detection of remote homologous proteins. By training on real structure similarities, the deep learning method captures subtle sequence similarities that indicate shared evolutionary origins.
About the Research
In the present paper, the authors proposed PLMSearch for homologous protein search driven by a protein language model. They designed this method to analyze protein sequences and identify evolutionary relationships based solely on sequence information. The study utilized deep representations from a pre-trained protein language model to train a similarity prediction model, which uncovers remote homology information concealed within the sequences.
The study focused on enhancing the sensitivity of detecting evolutionary relationships between proteins. PLMSearch improves the accuracy of detecting remote homology by following a three-step process.
- The first step involves protein family clan (PfamClan) filtering, which is used to filter out protein pairs that share the Pfam clan domain. This step helps narrow down the search space and focus on proteins that are more likely to have evolutionary relationships.
- The second step is structural similarity prediction. PLMSearch utilizes the similarity prediction model, which is trained using the deep representations from the pre-trained protein language model. This model can accurately predict the similarity between query-target pairs based on their sequences. By leveraging the information captured by the protein language model, the newly developed method detects remote homology even when the sequences are not highly similar.
- Finally, in the third step, the predicted similarities are used to sort the search results. This sorting process ensures that the most relevant and similar protein sequences are presented first, searching results more efficient and accurate.
The authors also mentioned two related methods, PML alignment (PLMAlign) and PLM-basic local alignment search too (PLM-BLAST). PLMAlign is used to align protein pairs retrieved by PLMSearch and obtain alignment scores. This step helps to further validate the predicted similarities and refine the search results. On the other hand, PLM-BLAST is a variant of the popular BLAST algorithm that incorporates the protein language model. It is used as a baseline method for comparison with PLMSearch.
Research Findings
The experimental results of PLMSearch demonstrated its exceptional performance in searching millions of query-target protein pairs within seconds, similar to any-against-many sequence searching (MMseqs2). It not only provided fast search capabilities but also significantly increased sensitivity by over threefold. This meant that PLMSearch was highly effective in detecting evolutionary relationships between proteins, even when the sequences were dissimilar.
Furthermore, PLMSearch proved comparable to state-of-the-art structure search methods by excelling in recalling remote homology pairs with dissimilar sequences but similar structures. This was a significant achievement because traditional sequence search methods often struggled to identify such relationships. Despite differences in their sequences, PLMSearch's ability to uncover hidden similarities in protein structures highlighted its effectiveness in detecting evolutionary connections.
To validate the efficacy of PLMSearch, tests were conducted on two datasets: the structural classification of proteins-extended database 40 test (SCOPe40-test) and the Swiss-Prot protein knowledgebase (Swiss-Prot). The outcomes of these tests further confirmed PLMSearch's superior performance, consistently outperforming other methods in identifying hidden sequence similarities indicative of evolutionary connections and accurately detecting relationships between proteins that may have evolved from a common ancestor.
Applications
PLMSearch has applications in various areas within bioinformatics and structural biology. By using PLMSearch, researchers can gain insights into the functions of proteins, their three-dimensional structures, and how they have evolved. This information is crucial for understanding the mechanisms underlying biological processes and can have implications in various fields, including drug discovery, protein engineering, and evolutionary biology.
Furthermore, PLMSearch offers enhanced precision and speed in analyzing protein data. It can efficiently search through millions of query-target protein pairs within seconds, making it a valuable tool for large-scale analyses. The speed and accuracy of PLMSearch enables researchers to process vast amounts of data and obtain meaningful results on time.
Conclusion
In summary, the novel approach proved effective and robust in the field of homologous protein search. Its ability to accurately identify remote homology pairs with distinct sequences but similar structures opens new avenues for studying protein evolution and function.
Moving forward, the researchers acknowledged the limitations and challenges and suggested further refining the PLMSearch method to enhance its performance in detecting remote homologous proteins with even greater sensitivity and accuracy. They proposed investigating the applicability of PLMSearch in diverse biological contexts and expanding its utility in studying complex protein relationships and evolutionary patterns. Moreover, the authors highlighted the potential for integrating additional data sources and refining the model architecture to further improve the method's capabilities in homologous protein search.